Promethium Tutorials
Calculate Conformers from SMILES: Nemorexant Example Using Default Settings
1
Step 1
After logging into your Promethium account, on top navigation bar, click on "Workflows" -> "Conformer Search".
2
Step 2
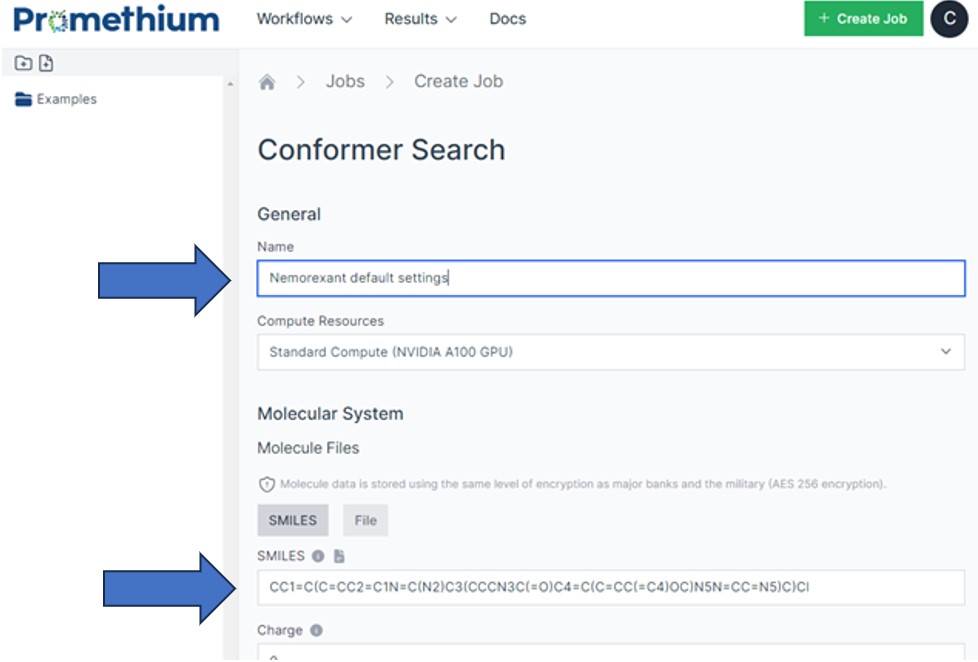
On the Conformer page:
-
Add a name for the job: e.g. "Nemorexant with default settings"
-
Add the following SMILES string to the SMILES input field: CC1=C(C=CC2=C1N=C(N2)C3(CCCN3C(=O)C4=C(C=CC(=C4)OC)N5N=CC=N5)C)Cl
3
Step 3
On bottom right, click "Review" and subsequently "Submit"
What happens now?
By default, there are 4 customizable filtering stages. Initial conformers are generated from stochastic sampling of the torsion angles.
- Stage 1: The initial conformers are optimized with a molecular mechanics force field.
- Stage 2: The lowest energy conformers are re-optimized using the ANI neural network potential.
- Stage 3: The lowest energy conformers are re-optimized at the B3LYP-D3/def2-SVP level of theory.
- Stage 4 (optional): The energy of each conformer is recalculated at the ωB97M-V/def2-TZVP level of theory.
.png?width=1040&height=821&name=image%20(13).png)
